Antiestrogens: structure-activity relationships and use in breast cancer treatment
- 1Institute for Research in Immunology and Cancer, Université de Montréal, Montréal, Québec, Canada
- 2Department of Biochemistry and Molecular Medicine, Université de Montréal, Montréal, Québec, Canada
- 3Department of Chemistry, McGill University, Montréal, Québec, Canada
- Correspondence should be addressed to S Mader; Email: sylvie.mader{at}umontreal.ca
-
Figure 1
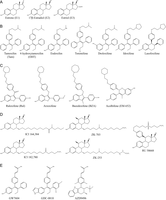
Chemical structures of estrogen receptor agonists and antagonists. (A) The three most abundant circulating estrogens: estrone, 17β-estradiol and estriol. (B) Tamoxifen and its active derivatives, 4-hydroxytamoxifen and endoxifen, as well as tamoxifen-derived SERMs. (C) Antiestrogens with different steroid-like backbones and a side chain containing a tertiary amine: SERM raloxifene and related compounds arzoxifene and bazedoxifene, as well as acolbifene. (D) Pure antiestrogens with steroid backbones and long side chains. (E) SERDs with steroid-like backbones and a side chain containing an acrylic acid.
-
Figure 2
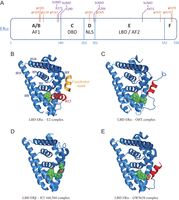
ERα structure, post-translational modifications and conformational changes induced by different ligands. (A) Schematic representation of ERα structure. AF1/AF2: activation function 1/2; DBD: DNA-Binding Domain; NLS: Nuclear Localization Signal; LBD: Ligand-Binding Domain. SUMOylation sites identified by mass spectrometry in the presence of ICI 182,780 are indicated in purple. Residues phosphorylated in the presence of antiestrogens or implicated in the modulation of sensitivity to antiestrogen treatment are indicated in orange. (B) LBD ERα – estradiol (E2) – TIF2 NR box 3 complex (Warnmark et al. 2002); (C) LBD ERα – 4-hydroxytamoxifen complex (OHT) (Shiau et al. 1998); (D) LBD ERβ – ICI 164,384 complex (Pike et al. 2001); (E) LBD ERα – GW5638 complex (Wu et al. 2005). Representations were generated using PyMOL. Helix 12 is highlighted in red and each ligand is shown in green. The α-helical TIF2 coactivator motif is shown in gold.
-
Figure 3
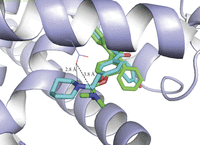
Role of Asp351 in the different activity of tamoxifen and raloxifene. Overlay of X-ray structures of 4-hydroxytamoxifen (green) and raloxifene (aqua) bound to ERα (data from Shiau et al. (1998) and Brzozowski et al. (1997), respectively). The distance from Asp351 to the dimethylamine in 4-hydroxytamoxifen (3.8 Å) is 1.0 Å longer than to the piperidine in raloxifene.
-
Figure 4
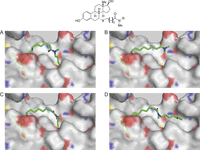
Models of ICI 164,384 and derivatives bound to ERβ. (A) ICI 164,384 (X = CH2, n = 9, R = C4H9); (B) a 13-atom side chain (X = S, n = 8, R = CH3); (C) a 15-atom side chain (X = S, n = 8, R = C3H7); (D) a 19-atom side chain (X = S, n = 8, R = C7H15); docking was performed using the Glide software as previously described (Hilmi et al. 2012).
- © 2017 The authors