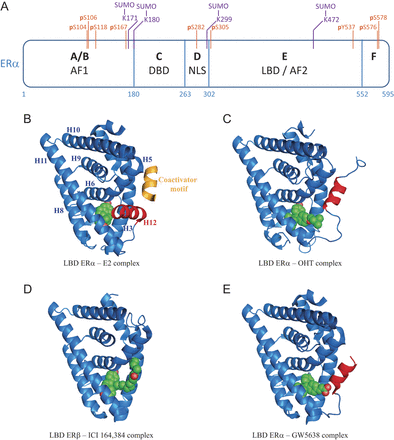
ERα structure, post-translational modifications and conformational changes induced by different ligands. (A) Schematic representation of ERα structure. AF1/AF2: activation function 1/2; DBD: DNA-Binding Domain; NLS: Nuclear Localization Signal; LBD: Ligand-Binding Domain. SUMOylation sites identified by mass spectrometry in the presence of ICI 182,780 are indicated in purple. Residues phosphorylated in the presence of antiestrogens or implicated in the modulation of sensitivity to antiestrogen treatment are indicated in orange. (B) LBD ERα – estradiol (E2) – TIF2 NR box 3 complex (Warnmark et al. 2002); (C) LBD ERα – 4-hydroxytamoxifen complex (OHT) (Shiau et al. 1998); (D) LBD ERβ – ICI 164,384 complex (Pike et al. 2001); (E) LBD ERα – GW5638 complex (Wu et al. 2005). Representations were generated using PyMOL. Helix 12 is highlighted in red and each ligand is shown in green. The α-helical TIF2 coactivator motif is shown in gold.











