60 YEARS OF POMC: Melanocortin receptors: evolution of ligand selectivity for melanocortin peptides
- Correspondence should be addressed to R M Dores; Email: rdores{at}du.edu
-
Figure 1
Proposed co-evolution of POMC and the MCR. This evolutionary tree for the chordates presents a working hypothesis for the origin of POMC and the MCRs. In this figure, proteins in white boxes etched with dashed lines are hypothetical, whereas proteins in colored boxes (green for POMC; blue or red for the MCRs) bordered by solid lines have been characterized in at least one species from the taxonomic group. For the MCRs, a blue box indicates a receptor that can be activated by ACTH or α-MSH, and a red box indicates a receptor that can only be activated by ACTH.The operating assumptions for this proposed scheme are that (a) the genes that code for the ancestral opioid precursor and the ancestral MCR emerged in a lineage of ancestral protochordates; (b) the first genome duplication event (1R) resulted in a gene that coded for POMC and a gene that coded for proenkephalin/prodynorphin precursor, and two genes that coded for paralogous MCRs (MCR′ and MCR″); (c) the second genome duplication event (2R) resulted in a gene that coded for POMC and four paralogous MCR genes; (d) a local gene duplication in the genome of the ancestral gnathostomes results in a fifth paralogous MCR gene. Among the extant cartilaginous fishes, all of the paralogous MCRs can be activated by ACTH or α-MSH (solid blue). Among the extant teleosts and tetrapods, only the MC2R (solid red) has exclusive selectivity for ACTH.1R, first chordate genome duplication event; 2R, second chordate genome duplication event; POMC, common precursor for ACTH and the MSHs; Proenk/Prodyn, a hypothetical protein that would have been ancestral to the opioid precursors proenkephalin and prodynorphin; MC*, ancestral MCR; MC′ and MC″, predicted hypothetical MCR paralogs after the 1R duplication event; MC1R, melanocortin-1 receptor; MC2R, melanocortin-2 receptor; MC3R, melanocortin-3 receptor; MC4R, melanocortin-4 receptor; MC5R, melanocortin-5 receptor.
-
Figure 2
Organization of POMC and comparison of vertebrate ACTH sequences. (A) The diagram provides a schematic representation of the positioning of chemical signals within the precursor protein, POMC. ACTH (red box) is positioned in roughly the middle of POMC. The first 13 amino acids in ACTH (marked off by the dashed line) are the sequence of α-MSH. The β-MSH sequence (blue box) is N-terminal to the sequence for the opioid, β-endorphin (black box). The γ-MSH sequence (white box, light green dash border) is present in cartilaginous fish and tetrapod POMC sequences (Dores & Baron 2011). The δ-MSH sequence (white box, dark green dash border) is only found in the POMC sequences of cartilaginous fishes (Takahashi et al. 2001). (B) The amino acid sequences of the following ACTH sequences were aligned: dogfish (Squalus acanthias; accession number AAS6720.1), Carp (Cyprinus carpio; accession number CAA74968.1), lungfish (Neoceratodus forsteri; accession number AAD37347.1), xenopus (Xenopus laevis; accession number AAH54160.1), bovine (Bos taurus; accession number NP_776576.1). Amino acid positions that were identical in four or more of the sequences are in red.
-
Figure 3
Comparison of human MC4R and MC2R. (A) In this schematic representation, the transmembrane domains in the human hMC4R are represented by the spheres and are numbered 1 through 7. The red spheres form the HFRW binding site for α-MSH (Pogozheva et al. 2005). The residues required for facilitating the binding of α-MSH are listed (hMC4R accession number – NP-005903.2). (B) The amino acid sequence of the human hMC2R is presented (accession number – NP-001278840.1). Amino acid positions that are also found in the HFRW binding site for hMC4R (Pogozheva et al. 2005) are marked with asterisk (*). Amino acid positions where a single alanine substitution decreased the activation of the mutant receptor expressed in OS3 adrenal cells are shaded in blue (Chen et al. 2007). Amino acid positions where amino acid substitutions as a result of a spontaneous point mutation that resulted in type 1 FGD and blocked activation of the mutant receptor (Chung et al. 2008) are shaded in red. The black bar demarcates the amino acid positions where single alanine mutations were made for the experiments presented in Figs 4 and 5. (C) A schematic representation of MC2R. The transmembrane domains are numbered as in Fig. 3A. The putative HFRW binding site for ACTH is represented by the transmembrane domains shaded in red. The putative KKRRP binding site for ACTH is represented by extracellular loop 2 (EC2) and transmembrane domain 5 shaded in green. The critical residues in this region that affect activation are listed based on the results from Figs 4 and 5 (data from Reinick et al. 2012b).
-
Figure 4
Analysis of single alanine mutants of hMC2R – TM4. For these experiments, hMC2R was synthesized with a V-5 epitope tag, and the mouse MRAP1 (accession number – NM_029844.3) was synthesized with a FLAG epitope tag by GenScript (Piscataway, NJ, USA). Both cDNA constructs were individually inserted in a pcDNA3.1+ vector for transfection into CHO cells. In addition, single alanine mutants of hMC2R were also made by GenScript, and the mutant cDNAs were individually co-transfected with mMRAP1 in CHO cells. The list of single alanine mutants is presented in panel A. CHO cells were transfected with the following plasmids: either human MC2R or human single alanine mutant MC2R, mouse mrap1, and the cAMP reporter plasmid cre-luc (Chepurny & Holz 2007) using a Cell Line Nucleofector Kit (Amaxa, Inc., Allendale, NJ, USA; www.lonza.com) as described by Liang et al. (2011). After 48 h, the transfected cells were stimulated with hACTH(1–24) (New England Peptide, Gardner, MA, USA) in serum-free CHO media for 4 h at 37°C at concentrations ranging from 10–7 to 10–13 M. After a 4-h incubation period, 100 µL of Bright-Glo Luciferase Assay System (Promega) was applied to each well and incubated at room temperature for 5 min. Luminescence was then measured using a BioTek Synergy HT plate reader. Average values and standard errors of the mean were graphed using KaleidaGraph software (v 4.1; www.synergy.com), and the EC50 value for each ligand was determined (A). The EC50 value for the wt MC2R/mMRAP1 transfection was compared with the EC50 value for each mutant MC2R/mMRAP1 using the Student’s t-test (N=3). (A) This figure has the EC50 values for the 22 single alanine mutant MC2R constructed tested. Single asterisk (*) indicates a drop in the EC50 value of the mutant MC2R relative to the positive control with a P value between 0.04 and 0.01. Double asterisks (**) indicate a drop in EC50 values of the mutant MC2R relative to the positive control with a P value <0.01. (B) The dose–response curves for the transmembrane 4 (TM4) mutant constructs are presented.
-
Figure 5
Analysis of single alanine mutants of hMC2R – EC2 and TM5. The activation assays were done as described in the legend for Fig. 4. (A) Presentation of the dose–response curves for extracellular loop 2 (EC2) mutant constructs. (B) Presentation of the dose–response curves for the transmembrane 5 (TM5) mutant constructs.
-
Figure 6
Interaction between MRAP1 and MC2R. This schematic presentation of the MC2R/MRAP1 heterodimer is based on data from the study by Cooray et al. (2011), with some modifications as a result of the study by Malik et al. (2015). MC2R (red sphere) will form a homodimer with two MRAP1 homodimers. In the MRAP1 homodimer, the N-terminal domain of each monomer is capped by a black triangle. The green box in the N-terminal domain is the activation motif. The purple zone on each monomer is the transmembrane domain. A star marks the activation motif that will make contact with an extracellular domain on MC2R.
-
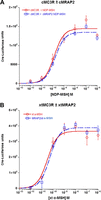
Figure 7
The effect of MRAP2 on the activation of chicken and frog MC3R. (A) A chicken (Gallus gallus) mc3r (cMC3R; accession number – BAA32555.1) cDNA was made by GenScript, and the cDNA was inserted into the pcDNA3.1+ expression vector. The cmc3r cDNA was expressed alone or co-expresses with a chicken mrap2 cDNA (cMRAP2; accession number – XP_004940463) in CHO cells as described in the legend for Fig. 4. All cells were transfected with the cre-luc cAMP reporter construct (Chepurny & Holz 2007). The transfected cells were stimulated with NDP-MSH (New England Peptide) at concentrations ranging from 10–12 to 10–6 M. Data are presented as mean±s.e.m. (N=3). The EC50 value for cMC3R was 1.4×10–10 M±5.4×10–11, and the EC50 value for cMC3R/cMRAP2 was 7.6 × 10–10 M±4.7×10–11. (B) A frog (Xenopus tropicalis) mc3r cDNA (xtMC3R; accession number – XP_002935936) was made by GenScript, and this cDNA was expressed either in the presence or absence of X. tropicalis mrap2 cDNA (xtMRAP2; accession number – XP_00293393) in CHO cells and stimulated with xt α-MSH (New England Peptide) at concentrations ranging from 10–12 to 10–6 M. Data are presented as mean±s.e.m. (N=3). The EC50 value for xtMC3R was 1.1×10–11 M±2.5×10–11, and the EC50 value for xtMC3R/xtMRAP2 was 6.5×10–11 M±1.3×10–11. Note that in (A) and (B), co-expression with the species-specific MRAP2 ortholog has no effect on the dose–response curves for either MC3R ortholog.
- © 2016 Society for Endocrinology