Characterisation of the androgen regulation of glycine N-methyltransferase in prostate cancer cells
- Silvia Ottaviani,
- Greg N Brooke1,
- Ciara O'Hanlon-Brown,
- Jonathan Waxman,
- Simak Ali and
- Laki Buluwela
- Department of Surgery and Cancer, Imperial College London, Imperial Centre for Translational and Experimental Medicine, London W12 0NN, UK
1School of Biological Sciences, University of Essex, Colchester, UK
- Correspondence should be addressed to L Buluwela or Ali, Email: l.buluwela{at}imperial.ac.uk; simak.ali{at}imperial.ac.uk
-
Figure 1
Androgen up-regulates the expression of GNMT mRNA and protein in a time-dependent manner. TaqMan RT-PCR for GNMT (A), PSA (B), NDRG1 (C) and TMPRSS2 (D) was performed from RNA prepared from LNCaP cells cultured in RPMI lacking phenol red supplemented with 10% DSS for 72 h followed by the addition of either ethanol or 1 nM R1881 for 24, 48 and 72 h. Data were normalised to GAPDH levels. The expression of each gene in cells treated with ethanol for 24 h was set to one, with the expression level in the other conditions being shown relative to this. Results are shown as mean values of three replicates with error bars showing s.e.m. (E) LNCaP cells were treated for the indicated times, whole cell lysates were separated by SDS–PAGE and immunoblotted for GNMT and β-actin. The polypeptide corresponding to GNMT is arrowed and molecular weight markers are expressed in kDa. (F) Immunofluorescence staining of GNMT was performed in LNCaP cells treated for 48 h. The GNMT antibody was detected with Alexa Fluor 488-labelled secondary and TO-PRO-3 was used for counterstaining of nuclei. All images were acquired using a Zeiss LSM510 confocal microscope. Scale bar=10 μm.
-
Figure 2
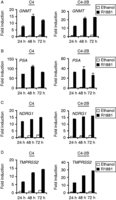
GNMT is androgen up-regulated in AR-positive cell lines. TaqMan RT-PCR for GNMT (A), PSA (B), NDRG1 (C) and TMPRSS2 (D) was performed using RNA prepared from LNCaP-derived lines C4 and C4-2B. Cells were cultured and treated using the same conditions described for LNCaP cells in Fig. 1.
-
Figure 3
AR knockdown suppresses the androgen-induced GNMT gene. (A) LNCaP cells were cultured in RPMI containing 10% DSS in the absence of antibiotics for 24 h. Cells were then transfected with siRNAs targeting AR or control. Twenty-four hours after transfection, cells were treated with 1 nM R1881 with an equal volume of ethanol being added to the vehicle control. RNA was prepared after a further 48 h and TaqMan RT-PCR for AR and GNMT was performed. Data have been normalised to GAPDH levels and AR or GNMT expression levels in the siRNA control treated with R1881 was set to one. Results are shown as mean values of three independent experiments performed in triplicates with error bars representing s.e.m. (B) LNCaP cells were transfected with siRNAs as for A. Whole cell lysates were prepared and immunoblotted for AR, GNMT (arrowed) and β-actin. Molecular weight markers are expressed in kDa. Suppression of the androgen-regulated genes PSA (C) and NDRG1 (D) following silencing of AR is also shown.
-
Figure 4
Site-directed mutagenesis analysis of the three predicted AREs. (A) The wild-type GNMT promoter reporter construct was generated by cloning a 1.2 kb region of the GNMT gene into a pGL3-basic luciferase vector. This construct was used as a template for site-directed mutagenesis of ARE-I (ARE-I*), ARE-II (ARE-II*) and ARE-III (ARE-III*). The mutants were generated by replacing part of the ARE sequence with the MluI restriction site sequence (5′-acggct-3′). (B) The activity of the GNMT ARE mutants was tested using the luciferase reporter assay. Firefly luciferase activities were normalised for transfection efficiency against the Renilla luciferase activities. Results are shown as mean of three independent experiments, each performed in triplicate. Error bars represent the s.e.m. Statistical significance was calculated by unpaired two-tailed Student's t-test: **P<0.01.
-
Figure 5
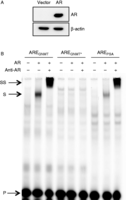
EMSA shows that AR binds to the ARE located within exon 1 of the GNMT gene. (A) COS-1 cells cultured in DMEM containing 10% FCS in the absence of antibiotics were transiently transfected with pSG5 control vector or AR expression plasmids. Twenty-four hours after transfection, cells were treated for 1 h with 10 nM R1881. High-salt buffer (HSB) cell extracts were prepared as described in the Materials and methods section. AR expression was confirmed by western blotting. (B) HSB cell extracts were first incubated with poly(dI.dC), in the absence or presence of an AR antibody, followed by incubation with the IR-800-labelled double-stranded oligonucleotides. The extract–oligonucleotide mix was resolved by electrophoresis through a 4% polyacrylamide gel and migration of the oligonucleotides through the gel was determined using the Odyssey (LI-COR) IR imaging system. Arrows indicate the positions of the unbound (P), shifted (S) and the antibody-AR-oligonucleotide ‘supershifted’ (SS) probe.
-
Figure 6
AR is recruited to the GNMT ARE in an androgen-dependent manner. (A) Diagramatic representation of the PSA (top) and GNMT (bottom) promoter regions. AREs are highlighted in boxes and arrows indicate the location of PCR primer pairs. (B) LNCaP cells were cultured in RPMI medium lacking phenol red supplemented with 10% DSS for 72 h followed by addition of 10 nM R1881 or ethanol for 1 h. Cells were then cross-linked, lysed and sonicated. Immunoprecipitations were performed using an antibody specific for AR or rabbit IgG. Immunoprecipitated DNA was reverse cross-linked and recovered by phenol/chloroform extraction. Real-time PCR was carried out using primers for the regions indicated in part A. The PSA enhancer region was chosen as a positive control, and a region distant from ARE elements as a negative control. Results show the mean values obtained from ChIP analysis of two replicates.
-
Figure 7
Nucleotide sequences of the region flanking the GNMT ARE in multiple species are shown. The box demarks the region of ARE homology and the translation start site is underlined. Sequences were retrieved from the UCSC (http://genome.ucsc.edu) using the following genome builds: hg19 (human), mm10 (mouse), rn5 (rat), oryCun2 (rabbit), anoCar2 (lizard), xenTro3 (xenopus) and danRer7 (zebrafish). Multiple-sequence alignment was performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Any nucleotide change from the human sequence is in bold and underlined. ‘*’ indicates positions that have a single, fully conserved residue; ’.’ indicates conservation between groups of weakly similar properties (scoring≤0.5 in the Gonnet PAM 250 matrix).
- © 2013 Society for Endocrinology