BRCA2 functions: from DNA repair to replication fork stabilization
- 1Laval University Cancer Research Center, CHU de Québec Research Center - Université Laval, Hôtel-Dieu de Québec, Oncology Axis, Quebec City, Canada
- 2Lady Davis Institute for Medical Research, Segal Cancer Centre, Jewish General Hospital, Montreal, Canada
- 3Department of Oncology, McGill University, Montreal, Canada
- Correspondence should be addressed to A Orthwein or A Fradet-Turcotte; Email: alexandre.orthwein{at}mcgill.ca or Amelie.Fradet-Turcotte{at}crchudequebec.ulaval.ca
-
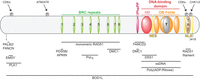
Figure 1
Structural domains and interaction partners of BRCA2. The N-terminal domain of BRCA2 is involved in several protein–protein interactions, including PALB2 and EMSY. BRCA2 contains eight BRC repeats located in the central portion of the protein; they are primarily involved in binding to monomeric RAD51, although they also are implicated in additional protein–protein interactions (PDS5B/APRIN and Polη). The BRCA2 DNA-binding domain (DBD) is composed of a helical domain (HD), three oligonucleotide/oligosaccharide-binding (OB) folds and a Tower domain (T). They promote BRCA2 binding to single-stranded DNA (ssDNA) and poly(ADP-Ribose). This domain also associates with DSS1. Adjacent to the DBD is a phenylalanine-proline-proline (PhePP) motif involved in the interaction with DMC1. This region is also implicated in the binding of FANCD2. The C-terminus of BRCA2 contains the TR2 domain, which interacts with RAD51 nucleofilaments. It also contains two distinct nuclear localization signals (NLSs) that are critical for BRCA2 nuclear localization. BRCA2 is posttranslationally modified by several cyclin-dependent (CDK, PLK1) and DNA damage-dependent (ATM/ATR, CHK1/2) kinases.
-
Figure 2
BRCA2 functions in the maintenance of genome stability. Bound to BRCA1 and PALB2, BRCA2 participates in multiple biological processes that are critical to maintain genome stability. First, BRCA2 is a key player in the repair of DNA lesions including DNA double-strand breaks (DSBs) and intrastrand crosslinks (ICLs). Moreover, BRCA2 has a DNA repair-independent function: it prevents nucleolytic degradation at stalled replication forks. Both of these functions are directly or indirectly involved in the maintenance of telomeres. BRCA2 is required for the processing of R-loops in collaboration with the TREX-2 complex. More recently, BRCA2 has been involved in mitophagy and the clearance of damaged mitochondria, thereby indirectly preserving genome stability.
-
Figure 3
Role of BRCA2 during DSB repair, ICL repair and stabilization of stalled replication forks. (A) DSB is first detected by the MRE11–RAD50–NBS1 (MRN) complex, which triggers a cascade of phosphorylation and ubiquitylation events (not shown) that promote the recruitment of BRCA1 and CtIP to the break (reviewed in Dantuma & van Attikum 2016). In S/G2 phase of the cell cycle, CtIP, along with the exonucleases Exo1 and DNA2-BLM, promotes extensive DNA end-resection, a step that commits cells to repair DSBs by homologous recombination (HR). Next, loading of RAD51 on the 3′-resected end by the concerted action of BRCA1/PALB2 and BRCA2 initiates homology search and the formation of a D-loop, a structure that results from the invasion of the homologous template by the RAD51-coated DNA strand. DNA synthesis and processing of the D-loop by synthesis-dependent strand annealing, gene conversion or break-induced replication repair complete this error-free DNA repair process. (B) Recognition and repair of ICLs is initiated when two replication forks converge at the lesion. The subsequent recruitment of the proteins of the Fanconi anemia (FA) core complex (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, FANCM) along with FANCT, FAAP100, MHF1, MHF2, FAAP20 FAAP24 and BRCA1 triggers monoubiquitylation of the heterodimer FANCI/FANCD2. Once activated, the heterodimer promotes nucleolytic incision at the converged replication forks and releases the ICL from one of the strands. The latter incision, also referred as the ‘unhooking’, is performed by a complex composed of the nuclease scaffold (SLX4) and the endonucleases ERCC4-ERCC1, MUS81-EME1 and FAN1. Depending on their structure, the parental DNA strands will be replicated by translesion synthesis (TLS) polymerases (REV1 or POLζ) or repaired by HR. (C) Following replication fork stalling, forks need to be protected from excessive resection. Although the exact molecular events that lead to their stabilization are still unclear, evidence support a role of BRCA1, BRCA2 and FANCD2 in promoting the loading of RAD51 at the fork, an event that is essential to protect the degradation of nascent strands by the nucleases MRE11 and DNA2. Whether RAD51 is loaded on ssDNA that arises on the parental strand or on the nascent strand is unknown (Models 1 and 2). The forks can be reprimed or restarted, a step that is orchestrated by the TLS polymerases. When submitted to sustained replication stress or when replication forks are unable to bypass roadblocks, forks collapse and the intervention of nucleases generates DSBs that are subsequently repaired by HR.
- © 2016 Society for Endocrinology